Notes
Foreword
This tool was developed in order to allow users to compare ORFomes themselves under their own understanding of stringency. The question of stringency is especially important for BLAST searches, where threshold values (e-value) are usually under evaluated, and when similarity may be low (e.g. paralogs).
Although a few bidirectional BLAST pipelines may be publicly available, they are usually implemented in PERL and designed for UNIX family environments. These issues make their transfer for desktop computers at least demanding.
After the bi-directional matching hits are subjected to further analyses, namely straight global alignment of nucleotide sequences, global alignment based on the products, calculation of molecular evolution rates, etc.
Tecnhical Specifications
A personal desktop computer running on a Microsoft Windows environment (for other systems minor changes are needed). A Pentium IV (or equivalent) processor with at least 512 MB of RAM makes the minimum requisites for running this package.The supplied file should be decompressed respecting the internal folder information (e.g. using free ZipGenius) before execution. To unzip the package a password is needed, please send an e-mail (jmfa@fct.unl.pt) in order to receive it. This will allow me to warn users of important changes and issues regarding this application.
Input Data
Input files should be placed in the 'input' folder.
CDS/ORF files should be supplied in FASTA/Pearson format, one for each ORFome. Two examples are supplied (target_cds.fasta and test_cds.fasta) and users should be warned about the need to avoid special characters in the annotation line.
These same files should be used to produce BLAST databases with suitable acronyms. The user may command this application to build them or may choose to build them. In the latter case the resulting files should be placed in the '/blast/db' folder.
(..\bin\formatdb -i <path\orfome filename> -p F -o T -n <blast database acronym> )
GO terms file (e.g. go_terms.tab) were obtained from SGD (http://downloads.yeastgenome.org/literature_curation/go_terms.tab) and updated versions should be downloaded regularly. For organisms other than fungi the user are encouraged to get hold of more adequate versions.
GO Slim mapping files should be provided with identical structure to the one available at SGD (http://downloads.yeastgenome.org/literature_curation/go_slim_mapping.tab). Any other file using the same column arrangement will be suitable. The reference field values must be the ones used for the reference ORFome.
A set of test files are supplied and the user encouraged to use them in a test run of this application.
Start Program
In the 'biDiBlast' folder locate and execute 'start.bat'.
Graphic User Interface
The application is started executing the 'start.bat' batch file in the main folder. This file should be edited to force special memory configurations or particular Java Virtual Machines.
The GUI is divided in two panels: a parameter value panel at left, and a execution control panel at right.
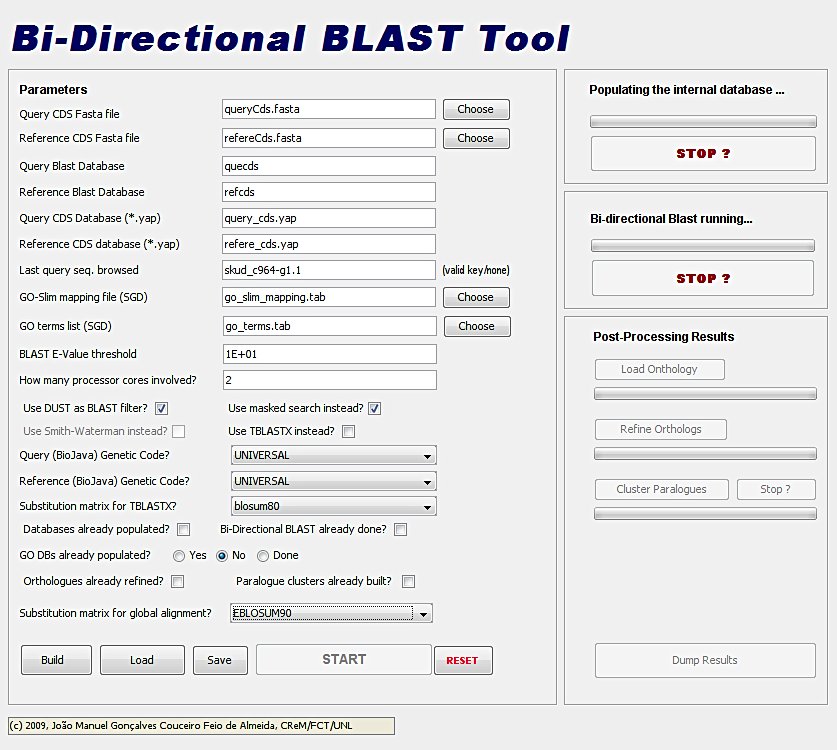
Starting the application the GUI appears with parameter boxes, and menus filled with values resulting from the last run. You can decide to keep them or change any value according to your needs, the result is saved pressing [Save]. After editing the parameters the [Build] button should be pressed unless the BLAST databases were already in place. The [START] button will always execute the 'next task'. The latter is a function of the flag parameters content that control the flow of the program:
- 'Databases already populated?'
- Checked - Uploads the ORF files into the databases (redundant entries are filtered)
- Unchecked - Assumes the databases are already populated
- 'Last query seq. browsed' - <accession nr.>/none
- <accession nr.> - Restarts the bi-directional BLAST comparisons from the referred entry
- none/no - This parameter value will be overlooked
- 'Bi-Directional BLAST already done?'
- Checked - In the next run the bi-directional BLAST comparison is assumed as done
- Unchecked - The comparisons are yet to be done
- 'GO DB's already populated?'
- done - The Slim GO objects were already created and linked to the putative homologue ORFs found
- yes - The GO-Slim objects were already created in the database
- no - The GO-Slim information is yet to be uploaded
- 'Orthologs already refined?'
- Checked - The sequence of the putative homologous ORFs and their conceptual translation were globally aligned (stretcher, EMBOSS). The substitution matrix employed is the 'SubstMat' at 'data' folder.
- Unchecked - Forces global alignment procedure
- 'Paralog clusters already built?'
- Checked - The putative paralogs were subjected to global alignment in the same terms as happened for the orthologs. The orthologues were used as reference for clustering paralogs.
- Unchecked - The procedure is yet to be executed
- 'Query CDS Fasta file' (../input)
- The name of the text file containing the uncharacterized CDSs
- 'Reference CDS Fasta file' (../input)
- The name of the text file containing the CDSs from the reference genome
- 'Query Blast Database'
- The acronym given to the BLAST database built upon the query CDSs. Avoid names with more than eight characters.
- 'Reference Blast Database'
- The acronym given to the BLAST database built upon the reference CDSs
- 'Query CDS Database (*.yap)'
- The name of the file to be given to the internal database (DB4O format) built upon the query CDSs. The name should be informative as this file can be used for different comparisons
- 'Reference CDS Database (*.yap)'
- The name of the file to be given to the internal database (DB4O format) built upon the reference CDSs
- 'GO-Slim mapping file (SGD)' (../input)
- The name given to a file with the same structure and data as http://downloads.yeastgenome.org/literature_curation/go_slim_mapping.tab
- 'GO terms list (SGD)' (../input)
- The name given to a file with the same structure and data as http://downloads.yeastgenome.org/literature_curation/go_terms.tab
- Query/Reference (BioJava) Genetic Code
- The genetic code for each set of coding sequences should be given in order to translate and align the conceptual products. The allowed values are defined through BioJava, and provided through a drop-down menu:
- UNIVERSAL
BACTERIAL
YEAST_MITOCHONDRIAL
VERTEBRATE_MITOCHONDRIAL
MOLD_MITOCHONDRIAL
INVERTEBRATE_MITOCHONDRIAL
ECHINODERM_MITOCHONDRIAL
ASCIDIAN_MITOCHONDRIAL
FLATWORM_MITOCHONDRIAL
CILIATE_NUCLEAR
EUPLOTID_NUCLEAR
ALTERNATIVE_YEAST_NUCLEAR
BLEPHARISMA_MACRONUCLEAR
The global alignments of conceptual products made by the stretcher (EMBOSS) tool use a given substitution matrix (e.g. EBLOSUM62 for general purpose alignments). To obtain meaningful results the user should select an adequate matrix from the available set. A drop-down menu is provided.
When comparing sequences from genomes of different genera TBLASTX is recommended. This option is available through a check box.
There are also options for using filters during BLAST/TBLASTX searches. Those are the standard filters present in NCBI-BLAST: DUST for nucleotide sequences, and SEG if TBLASTX is chosen. The option for masked searches activates the filter in spite of its check box being unmarked.
Buttons
Build - Builds the BLAST databases based on the supplied FASTA files, and with the stated names.
Load - Load the parameter values as stored in the ../input/costumize.properties file.
Save - Stores the actual parameter values as present in the GUI.
Start - When enabled allow the user to start the procedure with the parameter values as stored in the ../input/costumize.properties file. The procedure will run without further action until the bi-directional BLAST search is completed.
Load Ontology - Loads the GO Slim terms (../input/go_terms.tab file) into the aplication internal database, and map them to the reference sequences using the ../input/go_slim_mapping.tab file.
Refine Orthologs - Initiates the process of global alignment of all the bi-directional BLAST hits. Statistics, and evolution rates are derived at this stage.
Cluster Paralogs - The same procedure is carried on the uni-directional BLAST hits. After that these putative paralog sequences are clustered with orthologs with a common reference sequence.
Dump Results - Dumps the content of the application internal database into a set of text delimited tables. These are placed in the ../output folder.
Reset - Stores transient, and output files in compressed (zip) file in the ../output folder. The stored files are subsequently erased, and parameter values are reset. The user shoul press the Save button aftarwards.
Standard Procedure
Prior to the program execution all needed files should be placed in the 'input' and 'blast/db' folder.Start the program by calling 'start.bat'. Check the parameter values, press [Save] and wait. The progress in the upload of the internal databases and the execution of the bi-directional BLAST comparison is portrayed by the progress bars. At any time those processes could be interrupted if needed, pressing [STOP?].
After being through the comparisons it is advisable to save the resulting parameter set ([Save]). Press [Load] if the GUI does not refresh properly.
The next phase would be started by pressing [Load Ontology] and the other buttons down that panel.
Pressing [Dump Results] button will write the results to a set of files in the 'output' folder. These files may be imported into a relational database environment to be explored. At times this last button appears as inactive (toned down), but it may be pressed after the execution of each step. The amount of results to be dumped depend on the steps already executed.
Peculiarities
The graphic user interface (GUI) is still in development and from time to time it won't refresh properly. This problem has no impact on the underlying calculations or their results. Whenever the parameter fields are only refreshed the user may press [Load] button. After that it is advisable to press [Save] and then [START]. The program run will proceed according to the stated flags.The dumped results can be imported in a spreadsheet software although a relational database system is better suited for the analysis. The field (column) separator code is '»' to allow for complex annotation in the imported sequences. Inside the fields containing multiline content (e.g. alignment of sequences) '£' stands for <carriage return>.
The decimal separator character is '.'. Users using localization patterns with other decimal separators should consider the conversion in the import profiles.